About me
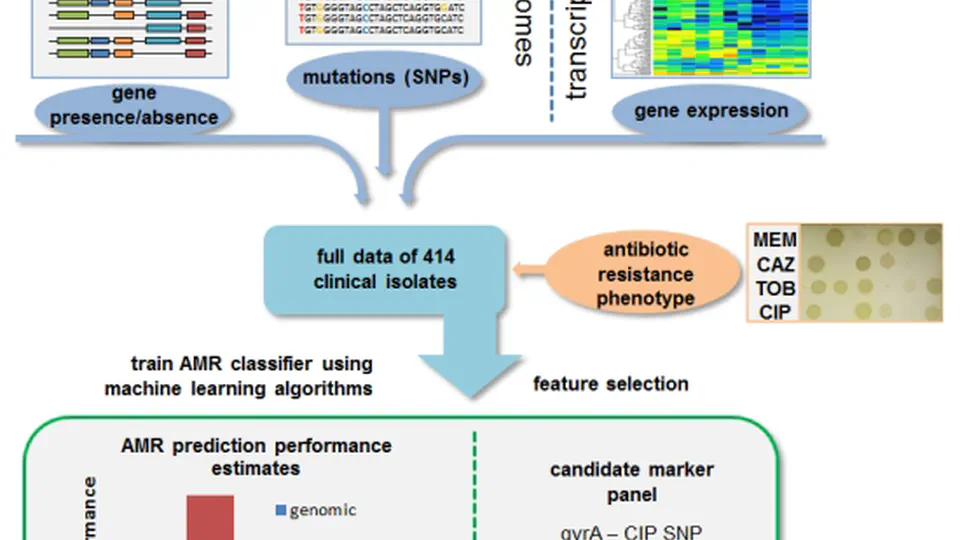
I am a postdoctoral scientist specializing in computational biology, with a strong focus on bacterial genomics. My expertise lies in developing and applying AI/ML-driven tools to analyse genomic patterns that influence bacterial pathogen behaviour, including antimicrobial resistance and virulence.
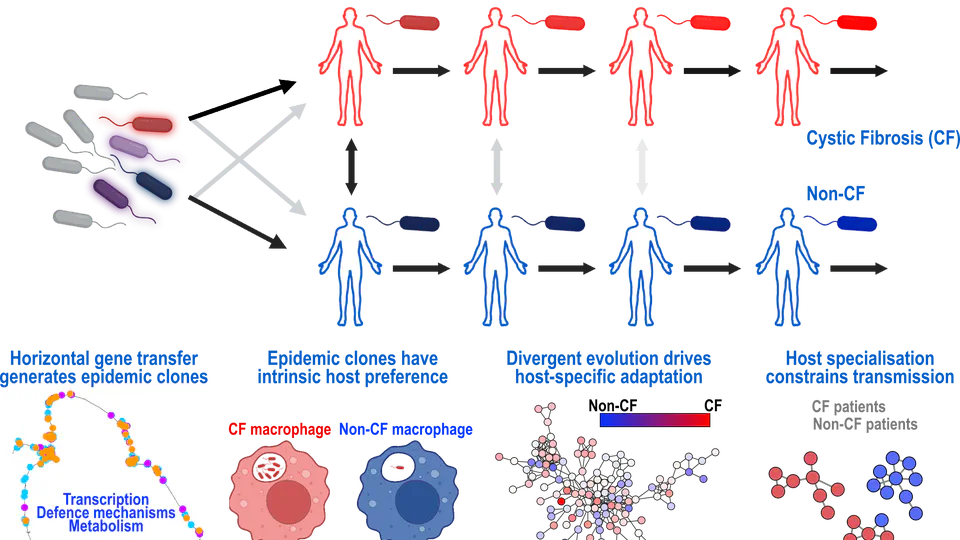
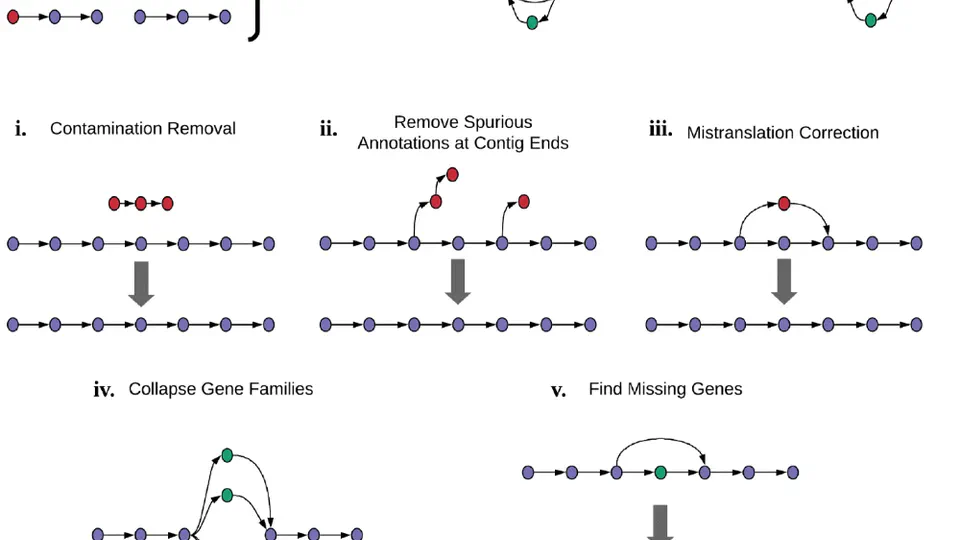
In recent years, my research has concentrated on Pseudomonas aeruginosa. Collaborating with academic and industry partners worldwide, we have put together and utilized the largest data collection globally to map the evolution of this bacterium. Our groundbreaking findings have revealed significant insights into how Pseudomonas aeruginosa has evolved and adapted to the human environment, with important implications for clinical practice.
- Microbiology
- Infectious diseases
- Evolutionary biology
- Phylogenetics
- Statistical genomics
- Machine learning and AI
PhD Computer Science, 2017
Heinrich Heine University of Duesseldorf
MSc Bioinformatics, 2010
Saarland University
BSc Bioinformatics, 2008
Saarland University
- Postdoctoral fellow, VPD Heart and Lung Research Institute, University of Cambridge with Professor Andres Floto
- Postdoctoral associate, Cambridge Center of AI in Medicine (CCAIM)
- Visiting postdoctoral fellow, University of Cambridge Veterinary School with Professor Julian Parkhill
- Visiting postdoctoral fellow, Wellcome Trust Sanger Institute with Professor Nicholas Thomson