Producing polished prokaryotic pangenomes with the Panaroo pipeline
Jul 1, 2020·,,,,,,,,,,,,,·
0 min read
Gerry Tonkin-Hill
Neil MacAlasdair
Christopher Ruis
Aaron Weimann
Gal Horesh
John a Lees
Rebecca a Gladstone
Stephanie Lo
Christopher Beaudoin
R Andres Floto
Simon D W Frost
Jukka Corander
Stephen D Bentley
Julian Parkhill
Abstract
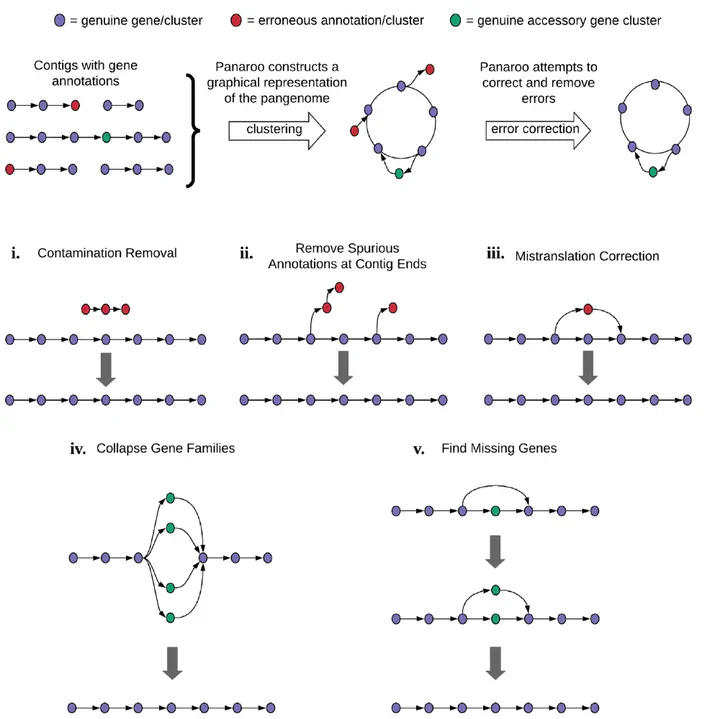
Population-level comparisons of prokaryotic genomes must take into account the substantial differences in gene content resulting from horizontal gene transfer, gene duplication and gene loss. However, the automated annotation of prokaryotic genomes is imperfect, and errors due to fragmented assemblies, contamination, diverse gene families and mis-assemblies accumulate over the population, leading to profound consequences when analysing the set of all genes found in a species. Here, we introduce Panaroo, a graph-based pangenome clustering tool that is able to account for many of the sources of error introduced during the annotation of prokaryotic genome assemblies. Panaroo is available at
.
Type
Publication
Genome Biol.