Evolution and host-specific adaptation of Pseudomonas aeruginosa
Jul 1, 2024·,,,,,,,,,,,,,,,,,,,,,,,,,,,,,·
0 min read
Aaron Weimann
Adam M Dinan
Christopher Ruis
Audrey Bernut
Stéphane Pont
Karen Brown
Judy Ryan
Lúcia Santos
Louise Ellison
Emem Ukor
Arun P Pandurangan
Sina Krokowski
Tom L Blundell
Martin Welch
Beth Blane
Kim Judge
Rachel Bousfield
Nicholas Brown
Josephine M Bryant
Irena Kukavica-Ibrulj
Giordano Rampioni
Livia Leoni
Patrick T Harrison
Sharon J Peacock
Nicholas R Thomson
Jeff Gauthier
Jo L Fothergill
Roger C Levesque
Julian Parkhill
R Andres Floto
Abstract
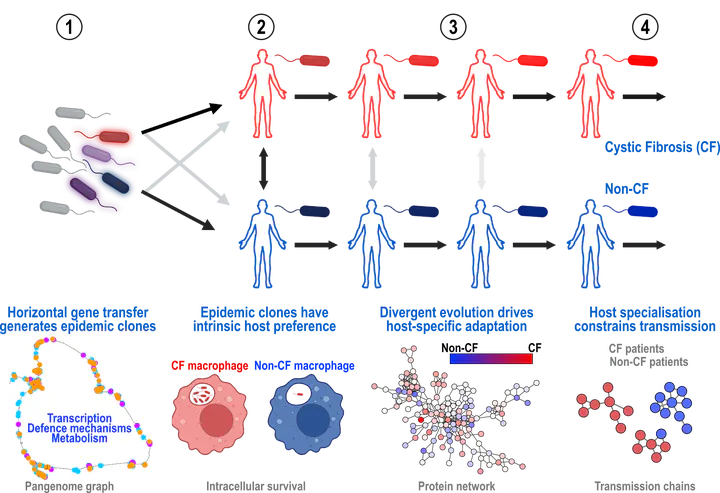
The major human bacterial pathogen Pseudomonas aeruginosa causes multidrug-resistant infections in people with underlying immunodeficiencies or structural lung diseases such as cystic fibrosis (CF). We show that a few environmental isolates, driven by horizontal gene acquisition, have become dominant epidemic clones that have sequentially emerged and spread through global transmission networks over the past 200 years. These clones demonstrate varying intrinsic propensities for infecting CF or non-CF individuals (linked to specific transcriptional changes enabling survival within macrophages); have undergone multiple rounds of convergent, host-specific adaptation; and have eventually lost their ability to transmit between different patient groups. Our findings thus explain the pathogenic evolution of P. aeruginosa and highlight the importance of global surveillance and cross-infection prevention in averting the emergence of future epidemic clones. Individuals with cystic fibrosis and other chronic inflammatory lung diseases are particularly vulnerable to colonization by Pseudomonas aeruginosa. Epidemic clones have emerged in certain patient groups that are associated with poor clinical outcomes and antimicrobial resistance. Weimann et al. investigated when and how such clones emerge by accessing international collections of clinical and environmental strains of P. aeruginosa amassed over the past century. The authors distinguished 21 major clones based on pairwise single-nucleotide polymorphism distances and majority multilocus sequence type. A series of epidemic clones emerged and expanded between 1850 and 2000, indicating recent selective pressures, possibly driven by industrialization, associated pollution, and human population change and underpinned by major changes in bacterial physiology. —Caroline Ash
Type
Publication
Science