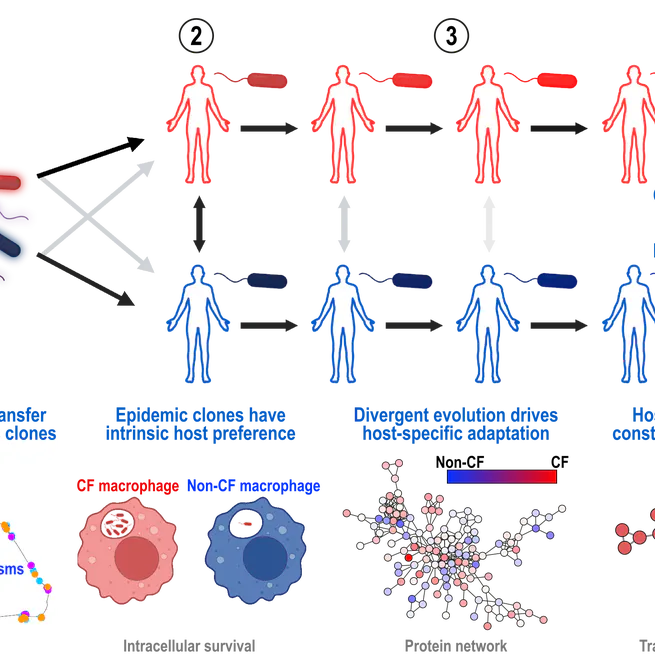
We describe how Pseudmonas aeruginosa has evolved over the last 200 years into a global pathogen via horizontal gene transfer and then further specialised to infect people with cystic fibrosis or without via convergent molecular evolution.
Jul 1, 2024
Nov 1, 2023
Nov 1, 2023
Sep 1, 2022
Apr 1, 2021
Jan 1, 2021
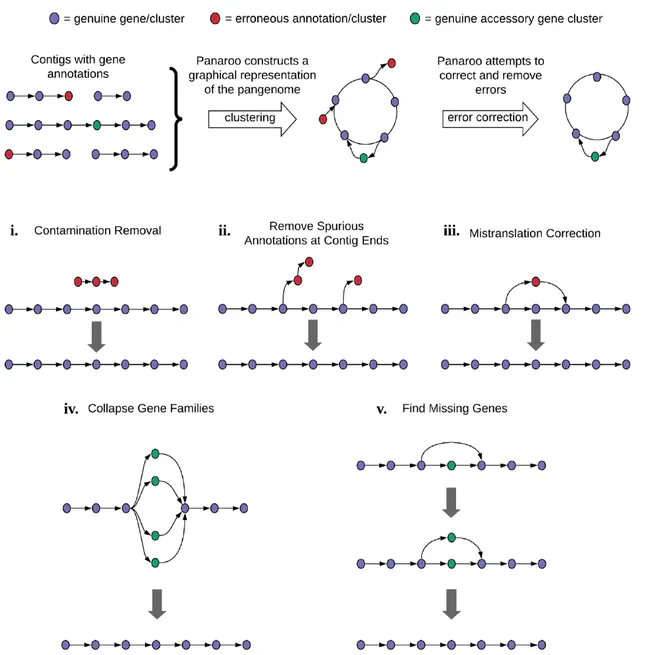
Panaroo is a a graph-based pangenome clustering tool that is able to account for many of the sources of error introduced during the annotation of prokaryotic genome assemblies. It's available at https://github.com/gtonkinhill/panaroo.
Jul 1, 2020
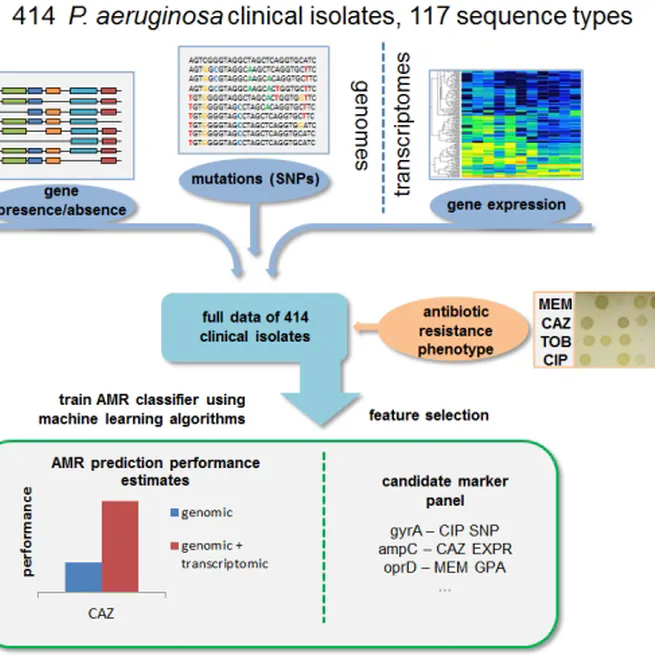
By training machine learning classifiers on information about the presence or absence of genes, their sequence variation, and expression profiles, we generated predictive models and identified biomarkers of resistance to four commonly administered antimicrobial drugs.
Mar 1, 2020
Nov 1, 2017
Jan 1, 2017